Project: High Angular Resolution Diffusion Imaging Tools
Example to illustrate the Spherical Deconvolution reconstruction via the Lucy-Richardson (L-R) algorithm. The fiber ODF is defined by the multi-tensor diffusion model.
- Language: MATLAB(R)
- Author: Erick Canales-Rodríguez, Lester Melie-García, Yasser Iturria-Medina, Yasser Alemán-Gómez
- Date: 2013, Version: 1.2
Contents
- Parameters for reconstruction: All this part is equal for each voxel
clear all; close all; clc; % - Loading the measurement scheme grad = load('/sampling_and_reconstruction_schemes/On_the_sphere/70_shell.txt'); % grad is a Nx3 matrix where grad(i,:) = [xi yi zi] and sqrt(xi^2 + yi^2 + zi^2) == 1 (unit vectors). display(['The measurement scheme contains ' num2str(size(grad,1)) ' directions']); % - Loading the reconstruction scheme (mesh where the fiber ODF will be reconstructed) % --------------------------------- % unit vectors on the sphere disp(' ') V = load('/sampling_and_reconstruction_schemes/On_the_sphere/724_shell.txt'); % facets display(['The reconstruction scheme contains ' num2str(size(V,1)) ' directions']); F = load('/sampling_and_reconstruction_schemes/On_the_sphere/724_sphere_facets.txt'); disp(' '); % - Creating the Kernel for the reconstruction. % --------------------------------- % Kernel is a Dictionary of diffusion tensor signals oriented along several % directions on the sphere (in our example V, which is the set of orientations where the fODF % will be computed). % It also includes two compartments with isotropic diffusion to fit real Brain data: CSF and GM % diffusivities assumed for the Dictionary lambda1 = 1.2*10^-3;% mm^2/s lambda2 = 0.0*10^-3;% mm^2/s (Like the ball and stick model of T. Behrens) % b-value and grad: equal to the experimental parameters used to adquire the data b = 3*10^3;% s/mm^2 grad = [0 0 0; grad]; % Add a measurement with b-value = 0 into the gradient matrix % (standard format in which the signal is measured in the scanner) SNR = 20; % This is a parameter required for the "create_signal_multi_tensor" function, % however, in this case the signal will be generated without noise. add_rician_noise = 0; [phi, theta] = cart2sph(V(:,1),V(:,2),V(:,3)); % set of directions Kernel = zeros(length(grad),length(V) ); S0 = 1; % The Dictionary is created under the assumption S0 = 1; fi = 1; % volume fraction for i=1:length(phi) anglesFi = [phi(i), theta(i)]*(180/pi); % in degrees Kernel(:,i) = create_signal_multi_tensor(anglesFi, fi, [lambda1, lambda2, lambda2], ... b, grad, S0, SNR, add_rician_noise); end % adding the isotropic compartment for CSF lambda1 = 2.0*10^-3;% mm^2/s Kernel(:,length(phi) + 1) = create_signal_multi_tensor([0 0], fi, [lambda1, lambda1, lambda1], ... b, grad, S0, SNR, add_rician_noise); % adding the isotropic compartment for GM lambda1 = 0.7*10^-3;% mm^2/s Kernel(:,length(phi) + 2) = create_signal_multi_tensor([0 0], fi, [lambda1, lambda1, lambda1], ... b, grad, S0, SNR, add_rician_noise); % ---------------------------------
The measurement scheme contains 70 directions The reconstruction scheme contains 724 directions
- Simulated diffusion MRI signal using a multi-tensor model
% --- Experimental parameters --- % % new real diffusivities lambda1 = 1.5*10^-3;% mm^2/s lambda2 = 0.3*10^-3;% mm^2/s % S0 is the image at b-value = 0 S0 = 100; % Signal to noise ratio SNR = 20; display(['The signal to noise ratio in this experiment is SNR = ' num2str(SNR) '.']); % --- Intravoxel properties --- % % Using a mixture of two fibers (more fibers can be used) % volume fraction for each fiber (the sum of this elemenst is 1 by definition) f1 = 0.5; f2 = 0.5; % angular orientation corresponding to each fiber (main eigenvector for each diffusion tensor) anglesF = [0 0; 55 0]; % [ang1 ang2]: -> ang1 is the rotation in degrees from x to y axis (rotation using the z-axis); % ang2 is the rotation in degrees from the x-y plane to the z axis (rotation using the y axis) % anglesF(i,:) defines the orientation of the fiber with volume fraction fi. % --- Computation of the diffusion signal using the above experimental parameters and intravoxel properties % % *** Signal with realistic rician noise add_rician_noise = 1; S = create_signal_multi_tensor(anglesF, [f1 f2],... [lambda1, lambda2, lambda2], b, grad, S0, SNR, add_rician_noise);
The signal to noise ratio in this experiment is SNR = 20.
- Initial estimate of the S0 component from the signal
% --- find zeros
grad_sum = sum(abs(grad),2);
ind_0 = find( grad_sum == 0);
S0_est = mean(S(ind_0));
- fODF estimation via the standard and damped L-R algorithms
% --- parameters definition %type_of_noise = 'Gaussian'; % (Default reconstruction published by Flavio Dell’Acqua!) type_of_noise = 'Poisson'; % (Experimental, not extensively tested, it provides sharper solutions) Niter = 200; % optimal value between 200 and 400 (for Gaussian noise) % -- Initial solution: Constant value on the sphere, non-negative iso-probable spherical function fODF0 = ones(size(Kernel,2)); fODF0 = fODF0/sum(fODF0); % --- standard L-R reconstruction display(' ') display('This is the real/effective time required for computing the fODF at each voxel') display(' via the standard L-R algorithm:') % tic [fODF_std, fgm1, fcsf1] = sph_deconv_RL(S/S0_est, Kernel, fODF0, Niter, type_of_noise); % fgm is the fraction of the isotropic component in GM tissues. % fcsf is the fraction of the isotropic component in CSF. toc % --- damped L-R reconstruction display(' ') display('This is the real/effective time required for computing the fODF at each voxel') display(' via the damped L-R algorithm:') % tic [fODF_d, fgm2, fcsf2] = sph_deconv_RLdamp(S/S0_est, Kernel, fODF0, Niter, type_of_noise); % fgm is the fraction of the isotropic component in GM tissues. % fcsf is the fraction of the isotropic component in CSF. toc
This is the real/effective time required for computing the fODF at each voxel via the standard L-R algorithm: Elapsed time is 0.027782 seconds. This is the real/effective time required for computing the fODF at each voxel via the damped L-R algorithm: Elapsed time is 0.069130 seconds.
- fODF estimation via non-negative least squares
display(' ') display('This is the real/effective time required for computing the fODF at each voxel') display(' via the linear least squares algorithm with nonnegativity constraints:') % options = optimset('lsqnonneg'); options = optimset(options,'TolX',1e-14); tic fODF_pos = lsqnonneg(Kernel, S/S0_est,options); fgm3 = fODF_pos(end); % fraction of the isotropic component for GM fcsf3 = fODF_pos(end-1); % fraction of the isotropic component for CSF fODF_pos(end-1:end) = []; % purely anisotropic part toc
This is the real/effective time required for computing the fODF at each voxel via the linear least squares algorithm with nonnegativity constraints: Elapsed time is 0.024180 seconds.
- Plots
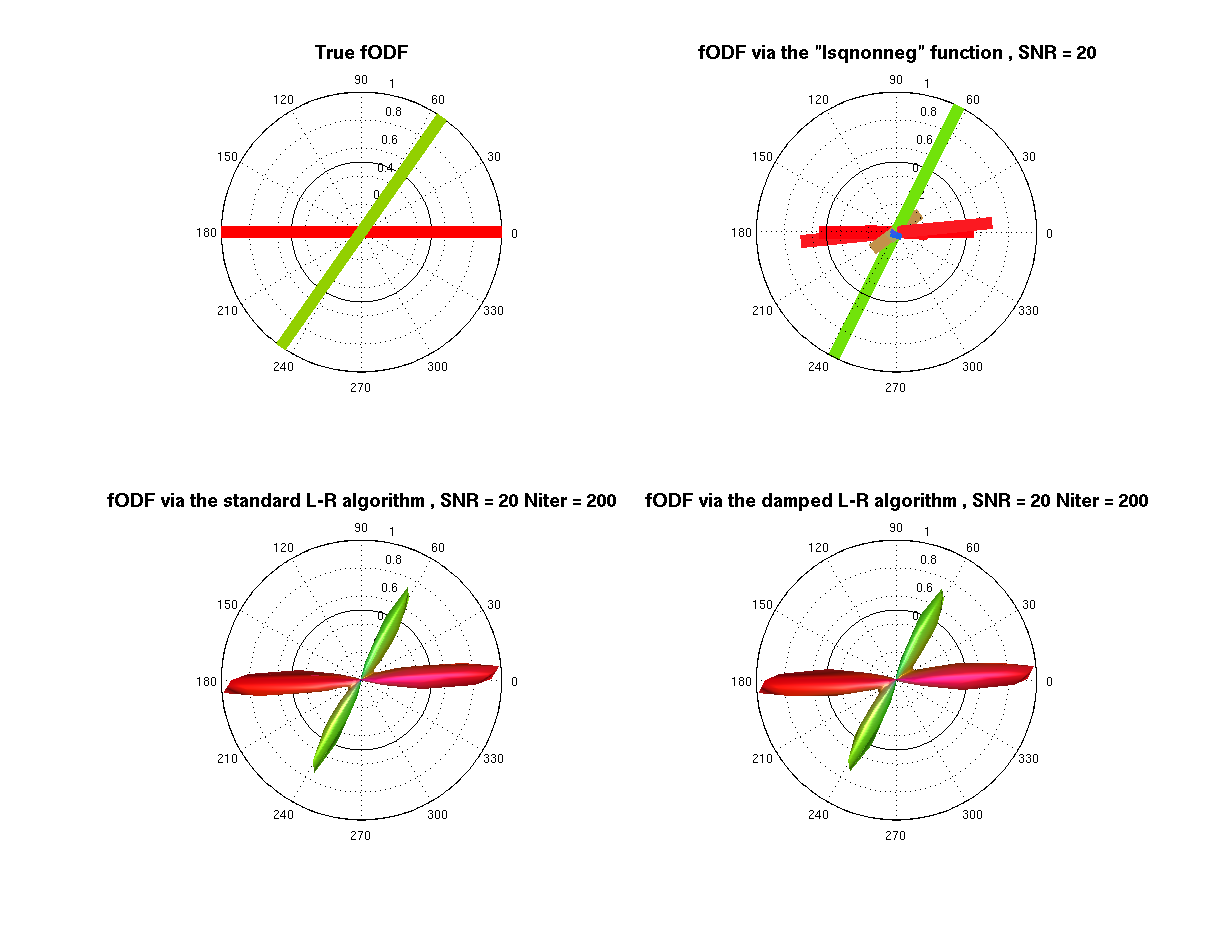
% --- opengl('software'); feature('UseMesaSoftwareOpenGL',1); set(gcf, 'color', 'white'); screen_size = get(0, 'ScreenSize'); f1 = figure(1); hold on; set(f1, 'Position', [0 0 screen_size(3) screen_size(4) ] ); cameramenu; title('Spherical deconvolution methods') % ----- Origen = [0 0 0.25]; % Voxel coordinates [x y z] where the ODF will be displayed angle = 0:.01:2*pi; R = ones(1,length(angle)); R1 = R.*f2/f1; subplot(2,2,1) polarm(angle,R,'k'); hold on; polarm(angle, R1,'k'); % --- anglesF = anglesF*(pi/180); [Vx, Vy, Vz] = sph2cart(anglesF(:,1), anglesF(:,2), ones(size(anglesF,1),1)); for i=1:size(Vx,1) hold on Color_i = [Vx(i) Vy(i) Vz(i)]/sqrt( Vx(i)^2 + Vy(i)^2 + Vz(i)^2 ); Color_i = Color_i.*sign(Color_i); line([-Vx(i) Vx(i)], [-Vy(i) Vy(i)], [-Vz(i) Vz(i)], 'LineWidth', 10, 'Color', Color_i); end % --- title('\fontsize{14} True fODF', 'FontWeight','bold') axis equal; axis off; subplot(2,2,2) polarm(angle,R,'k'); hold on; polarm(angle, R1,'k'); % --- % the fODF is scaled only for the display fODF_pos = fODF_pos./max(fODF_pos); ind0 = find(fODF_pos == 0); fODF_pos(ind0) = []; Vaux = V; Vaux(ind0,:) = []; Vx = Vaux(:,1); Vy = Vaux(:,2); Vz = Vaux(:,3); for i=1:size(Vx,1) hold on Color_i = [Vx(i) Vy(i) Vz(i)]/sqrt( Vx(i)^2 + Vy(i)^2 + Vz(i)^2 ); Color_i = Color_i.*sign(Color_i); line(fODF_pos(i)*[-Vx(i) Vx(i)], fODF_pos(i)*[-Vy(i) Vy(i)], fODF_pos(i)*[-Vz(i) Vz(i)], 'LineWidth', 10, 'Color', Color_i); end % --- title(['\fontsize{14} fODF via the "lsqnonneg" function , SNR = ' num2str(SNR) ], 'FontWeight','bold') axis equal; axis off; subplot(2,2,3) polarm(angle,R,'k'); hold on; polarm(angle, R1,'k'); % the fODF is scaled only for the display plot_ODF(fODF_std./max(fODF_std), V, F, Origen); title(['\fontsize{14} fODF via the standard L-R algorithm , SNR = ' num2str(SNR), ' Niter = ' num2str(Niter) ], 'FontWeight','bold') axis equal; axis off; subplot(2,2,4) polarm(angle,R,'k'); hold on; polarm(angle, R1,'k'); % the fODF is scaled only for the display plot_ODF(fODF_d./max(fODF_d),V, F, Origen); title(['\fontsize{14} fODF via the damped L-R algorithm , SNR = ' num2str(SNR), ' Niter = ' num2str(Niter) ], 'FontWeight','bold') axis equal; axis off;